About
ONTViSc (ONT-based Viral Screening for Biosecurity)
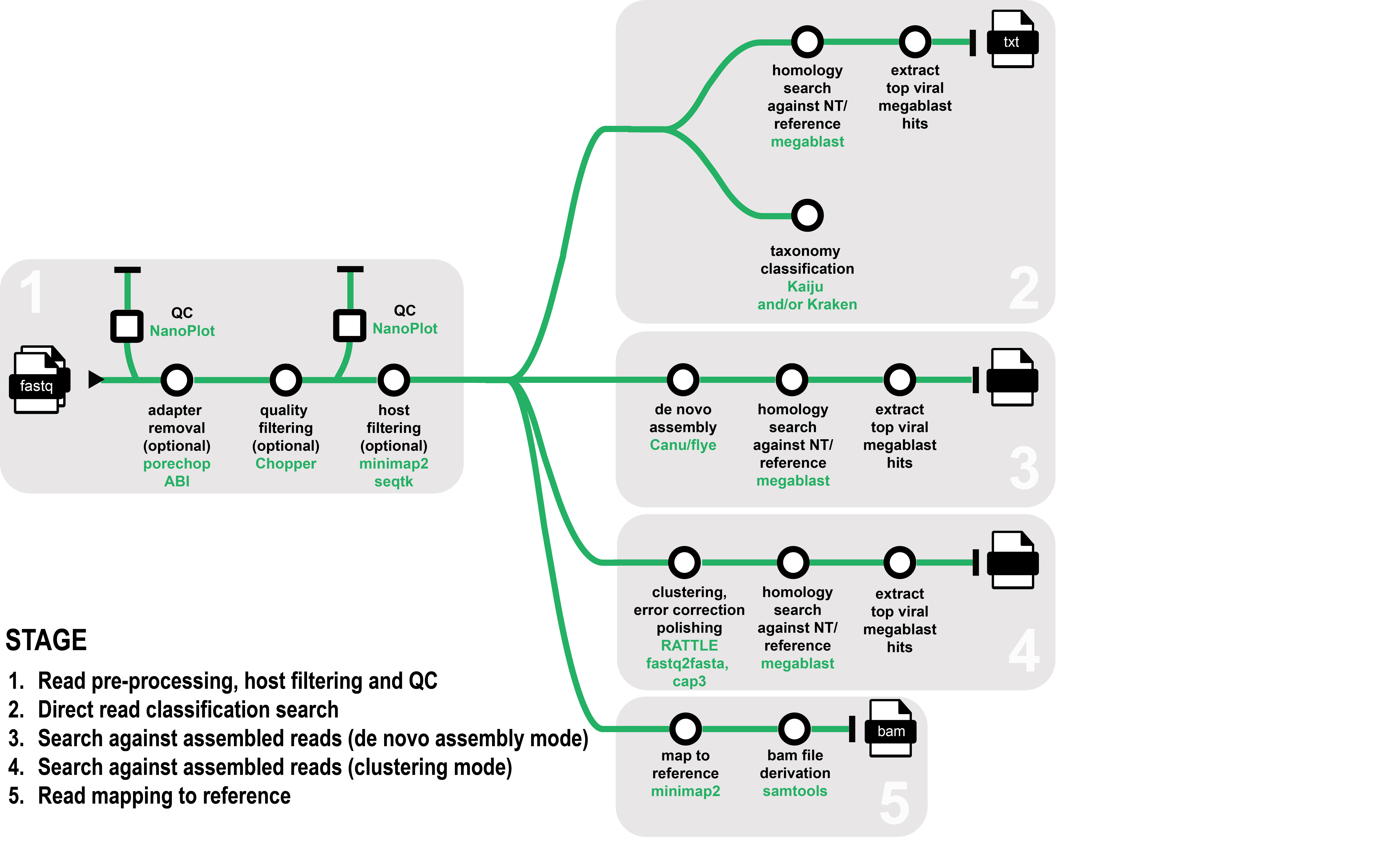
eresearchqut/ontvisc is a Nextflow-based bioinformatics pipeline designed to help diagnostics of viruses and viroid pathogens for biosecurity. It takes fastq files generated from either amplicon or whole-genome sequencing using Oxford Nanopore Technologies as input. The pipeline can either: 1) perform a direct search on the sequenced reads, 2) generate clusters, 3) assemble the reads to generate longer contigs or 4) directly map reads to a known reference. The reads can optionally be filtered from a plant host before performing downstream analysis.
This guide
In this guide draft, you will find instructions on how to set up and execute the ONTvisc pipeline on three high-performance computing systems: Lyra (QUT), Gadi (NCI) and Setonix (Pawsey).
Acknowledgements
This guide makes use of the ELIXIR toolkit theme

Thank you to Sarah Beecroft (Pawsey), Matthew Downton (Gadi) and Ziad Al-Bkhetan (Australian BioCommons) for help with testing the pipeline on Gadi and Setonix, as well as for providing valuable information regarding these two high-performance computing environments. Thank you also to the eResearch team from QUT for making it possible to execute the ONTvisc pipeline on Lyra and for all the provided support.